Automated Fruit Disease Detection and Grading Using Image Processing and Hybrid Deep Learning Models
DOI:
https://doi.org/10.26438/ijcse/v13i11.99107Keywords:
Convolutional Neural Networks (CNN), Fruit Disease Detection, Deep Learning, Image Processing, Agricultural Automation, Crop Health MonitoringAbstract
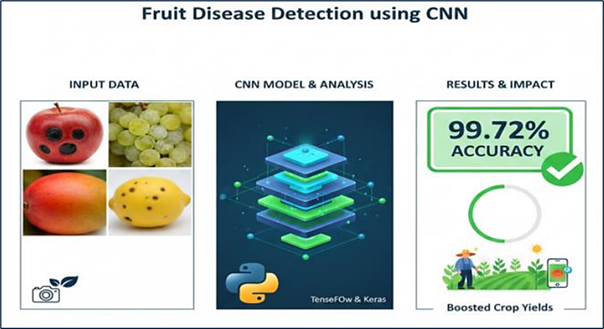
Automated fruit disease detection and grading are critical for advancing precision agriculture and reducing crop losses. This paper proposes a novel diagnostic framework that integrates advanced image processing techniques with a hybrid deep learning architecture. Two feature extraction pipelines were employed: statistical transforms, including Discrete Wavelet Transform (DWT) and Discrete Cosine Transform (DCT), and computer vision descriptors such as Scale-Invariant Feature Transform (SIFT), Speeded-Up Robust Features (SURF), and Gray-Level Co-occurrence Matrix (GLCM). The classification phase benchmarks five models—Decision Tree, K-Nearest Neighbor (KNN), Artificial Neural Network (ANN), Convolutional Neural Network (CNN), and a proposed Hybrid CNN+ANN architecture. Experimental results on a diverse fruit dataset demonstrate that the hybrid model achieves 99.54% accuracy with near-perfect precision and recall, outperforming traditional and standalone deep learning approaches. The system exhibits rapid convergence, scalability, and robustness against variations in lighting and background conditions. Integrated into a Python-based interface, this solution enables real-time disease diagnosis and grading, offering significant potential for smart farming and sustainable agricultural practices.
References
[1] M. A. Bhange and H. A. Hingoliwala, "A review of image processing for pomegranate disease detection," Int. J. Comput. Sci. Inf. Technol., vol. 6, no. 1, pp. 92–94, 2015.
[2] J. G. A. Barbedo, "Digital image processing techniques for detecting, quantifying and classifying plant diseases," SpringerPlus, vol. 2, no. 1, p. 660, Dec. 2013.
[3] P. Kaur and K. Singh, "Detection and classification of apple fruit diseases using image processing techniques: A review," J. Food Meas. Charact., vol. 14, no. 4, pp. 1364–1384, Aug. 2020.
[4] V. Kumar, R. Chhabra, and P. Singh, "Automatic detection and classification of apple diseases: A review," Comput. Electron. Agric., vol. 153, pp. 221–233, Oct. 2018.
[5] D. Unay and B. Gosselin, "A quality grading approach for `Jonagold` apples," in Proc. IEEE Benelux Signal Process. Symp. (SPS), 2004.
[6] V. Leemans and M. F. Destain, "A real-time grading method of apples based on features extracted from defects," J. Food Eng., vol. 61, no. 1, pp. 83–89, Jan. 2004.
[7] S. V. Gorawala, H. P. Patil, and A. R. Dixit, "Review of techniques for detection and classification of fruit diseases using image processing," Int. J. Comput. Appl., vol. 180, no. 12, pp. 7–11, 2018.
[8] A. Srivastava and S. Agarwal, "Fruit disease detection using image processing techniques: A review," Int. J. Recent Technol. Eng., vol. 8, no. 1S3, pp. 510–515, 2019.
[9] S. Bhardwaj, R. Jain, and S. Gupta, "A review of fruit detection and recognition techniques for fruit harvesting robots," J. Clean. Prod., vol. 195, pp. 48–62, Sep. 2018.
[10] S. S. Sannakki and S. C. Shiralashetti, "Image processing techniques for plant disease detection," Int. J. Comput. Sci. Mobile Comput., vol. 6, no. 6, pp. 171–177, 2017.
[11] T. Deshpande, S. Sengupta, and K. S. Raghuvanshi, "Grading & identification of disease in pomegranate leaf and fruit," Int. J. Comput. Sci. Inf. Technol., vol. 5, no. 3, pp. 4638–4645, 2014.
[12] S. Suvarnakanakaraddi, P. Liger, A. Gaonkar, M. Alagoudar, and A. Prakash, "Analysis and grading of pathogenic disease of chilli fruit using image processing," in Proc. Int. Conf. Adv. Eng. Technol., 2014, ISBN: 978-93-84209-06-3.
[13] J. D. Pujari, R. Yakkundimath, and A. S. Byadgi, "Statistical methods for quantitatively detecting fungal disease from fruits’ images," Int. J. Intell. Syst. Appl. Eng., vol. 1, no. 4, pp. 60–67, 2013.
[14] J. D. Pujari, R. Yakkundimath, and A. S. Byadgi, "Grading and classification of anthracnose fungal disease of fruits based on statistical texture features," Int. J. Adv. Sci. Technol., vol. 52, pp. 121–132, 2013.
[15] S. P. Mohanty, D. P. Hughes, and M. Salathé, "Using deep learning for image-based plant disease detection," Front. Plant Sci., vol. 7, p. 1419, Sep. 2016.
[16] X. Zhang, S. Zhang, and M. Guo, "A survey on deep learning for plant classification," Neurocomputing, vol. 300, pp. 83–99, Jul. 2018.
[17] Y. Liao, D. Cheng, L. Zhang, Q. Huang, and B. Yang, "Deep learning for plant disease diagnosis: A comprehensive review," Comput. Electron. Agric., vol. 183, p. 105019, Apr. 2021.
[18] M. Jhuria, A. Kumar, and R. Borse, "Image processing for smart farming: Detection of disease and fruit grading," in Proc. IEEE 2nd Int. Conf. Image Inf. Process. (ICIIP), 2013, pp. 521–526.
[19] D. Moshou, C. Bravo, J. S. West, H. A. McCartney, and H. Ramon, "Automatic detection of ‘yellow rust’ in wheat using reflectance measurements and neural networks," Comput. Electron. Agric., vol. 70, no. 1, pp. 58–65, Jan. 2010.
[20] A. Al-Hiary, S. Bani-Ahmad, M. Reyalat, M. Braik, and Z. AlRahamneh, "Fast and accurate detection and classification of plant leaf diseases," Int. J. Comput. Appl., vol. 17, no. 1, pp. 31–38, 2011.
[21] S. R. Dubey and A. S. Jalal, "Apple disease classification using color, texture and shape features from images," Signal Image Video Process., vol. 10, no. 5, pp. 819–826, Jul. 2016.
[22] K. P. Ferentinos, "Deep learning models for plant disease detection and diagnosis," Comput. Electron. Agric., vol. 145, pp. 311–318, Feb. 2018.
[23] B. Liu, Y. Zhang, D. He, and Y. Li, "Identification of apple leaf diseases based on deep convolutional neural networks," Symmetry, vol. 10, no. 1, p. 11, Jan. 2018.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors contributing to this journal agree to publish their articles under the Creative Commons Attribution 4.0 International License, allowing third parties to share their work (copy, distribute, transmit) and to adapt it, under the condition that the authors are given credit and that in the event of reuse or distribution, the terms of this license are made clear.




