Unveiling Model Superiority: A Comprehensive Analysis of Deep Learning Architectures for Robust Breast Cancer Prediction and Generalization
DOI:
https://doi.org/10.26438/ijcse/v13i4.114Keywords:
Breast Cancer, Deep Learning,, Ultrasound Imaging, Transfer Learning, DenseNet121, InceptionV3Abstract
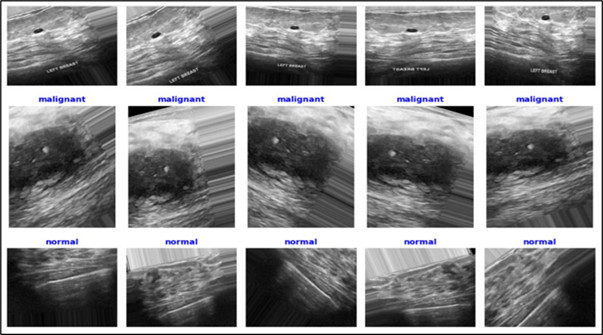
Breast cancer remains a leading global health challenge, demanding early, accurate, and interpretable diagnostic tools. This study presents a comprehensive evaluation of five pretrained convolutional neural networks—DenseNet121, InceptionV3, VGG19, EfficientNetB4, and MobileNetV3—for classifying breast ultrasound images from the BUSI dataset into Normal, Benign, and Malignant categories. The proposed framework integrates transfer learning, advanced preprocessing techniques, and class-weighted optimization to enhance model generalization and address data imbalance. Unlike prior studies, this work introduces a multi-model statistical comparison using Paired T-test, Wilcoxon Signed-Rank, and Cohen’s d, along with real-time inference benchmarking and a deployment-ready performance dashboard. Among the evaluated models, DenseNet121 demonstrated superior performance with an accuracy of 89.92% and an AUC-ROC of 0.95, outperforming existing state-of-the-art methods on the BUSI dataset. InceptionV3 also achieved strong results with 87.84% accuracy and notable inference speed. The findings confirm the clinical viability of integrating statistical rigor, inference-time awareness, and visual interpretability into deep learning pipelines for breast cancer detection. This framework lays the groundwork for scalable, explainable, and deployment-focused diagnostic AI systems in medical imaging.
References
[1] E. Du-Crow, "Computer-aided detection in mammography," M.Sc. thesis, Univ. of Manchester, 2022.
[2] A. Evans et al., "Breast ultrasound: Recommendations for information to women and referring physicians by the European Society of Breast Imaging," Insights Imaging, Vol.9, No.4, pp.449–461, 2018, doi: 10.1007/s13244-018-0637-8.
[3] G. Schueller, C. Schueller-Weidekamm, and T. H. Helbich, "Accuracy of ultrasound-guided, large-core needle breast biopsy," Eur. Radiol., Vol.18, No.8, pp.1761–1773, 2008. doi: 10.1007/s00330-008-0913-8.
[4] X. Shi, C. Liang, and H. Wang, "Multiview robust graph-based clustering for cancer subtype identification," IEEE/ACM Trans. Comput. Biol. Bioinf., Vol.20, No.2, pp.544–556, 2022, doi: 10.1109/TCBB.2022.3140846.
[5] H. Wang, G. Jiang, J. Peng, R. Deng, and X. Fu, "Towards adaptive consensus graph: Multi-view clustering via graph collaboration," IEEE Trans. Multimedia, Vol.24, pp.1–13, 2022, doi: 10.1109/TMM.2022.3142749.
[6] J. Bai, R. Posner, T. Wang, C. Yang, and S. Nabavi, "Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review," Med. Image Anal., vol. 71, p. 102049, 2021, doi: 10.1016/j.media.2021.102049.
[7] Z. Li, F. Liu, W. Yang, S. Peng, and J. Zhou, "A survey of convolutional neural networks: Analysis, applications, and prospects," IEEE Trans. Neural Netw. Learn. Syst., Vol.33, No.12, pp.6999–7019, 2022. doi: 10.1109/TNNLS.2022.3156148.
[8] J. Zuluaga-Gomez, Z. Al Masry, K. Benaggoune, S. Meraghni, and N. Zerhouni, "A CNN-based methodology for breast cancer diagnosis using thermal images," Comput. Methods Biomech. Biomed. Eng. Imag. Vis., Vol.9, No.2, pp.131–145, 2021, doi: 10.1080/21681163.2020.1796879.
[9] A. Krizhevsky, I. Sutskever, and G. E. Hinton, "ImageNet classification with deep convolutional neural networks," Commun. ACM, Vol.60, No.6, pp.84–90, 2017, doi: 10.1145/3065386.
[10] K. He, X. Zhang, S. Ren, and J. Sun, "Deep residual learning for image recognition," in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Las Vegas, NV, USA, pp.770–778, 2016. doi: 10.1109/CVPR.2016.90.
[11]B. Kaur and R. Popli, "A comprehensive review and analysis of advancements in breast cancer detection and classification," in Proc. 2024 3rd Edition of IEEE Delhi Section Flagship Conf. (DELCON), 2024, doi: 10.1109/DELCON64804.2024.10866898.
[12] A. M. Sharafaddini, K. K. Esfahani, and N. Mansouri, "Deep learning approaches to detect breast cancer: A comprehensive review," Multimedia Tools and Applications, Aug. 2024. https://doi.org/10.1007/s11042-024-XXXXX
[13] M. Mahalakshmi, G. R. Charan, and G. Sharma, "Integrative breast cancer detection: A deep learning approach with multi-modal data fusion of mammograms, prescription and blood reports," in Proc. 2024 Int. Conf. on Communication, Computing and Internet of Things (IC3IoT), 2024, doi: 10.1109/IC3IoT60841.2024.10550391.
[14] A. Alloqmani, Y. B. Abushark, and A. I. Khan, "Anomaly detection of breast cancer using deep learning," Arabian Journal for Science and Engineering, Vol.48, pp.10977–11002, 2023. doi: 10.1007/s13369-023-07914-2.
[15] S. S. Pandi, S. Anandhi, T. Kumaragurubaran, and B. Priyan, "Automatic breast cancer disease detection and diagnosis using learning algorithm," in Proc. 2024 2nd Int. Conf. on Advances in Information Technology (ICAIT), 2024. doi: 10.1109/ICAIT61638.2024.10690336.
[16] S. Garg, K. Kaur, N. Kumar, and J. J. P. C. Rodrigues, "Hybrid deep-learning-based anomaly detection scheme for suspicious flow detection in SDN: A social multimedia perspective," IEEE Trans. Multimedia, Vol.21, No.3, pp.566–578, 2019. doi: 10.1109/TMM.2019.2893549.
[17] O. Russakovsky et al., "ImageNet large scale visual recognition challenge," Int. J. Comput. Vis., Vol.115, No.3, pp.211–252, 2015. doi: 10.1007/s11263-015-0816-y.
[18] A. Krizhevsky, I. Sutskever, and G. E. Hinton, "ImageNet classification with deep convolutional neural networks," in Proc. 26th Conf. Neural Information Processing Systems (NIPS’12), Lake Tahoe, NV, USA, Dec., pp.1097–1105, 2012. doi: 10.1145/3065386.
[19] J. Arevalo, F. A. González, R. Ramos-Pollán, J. L. Oliveira, and M. A. G. Lopez, "Representation learning for mammography mass lesion classification with convolutional neural networks," Comput. Methods Programs Biomed., Vol.127, pp.248–257, 2016. doi: 10.1016/j.cmpb.2015.12.014.
[20] B. Q. Huynh, H. Li, and M. L. Giger, "Digital mammographic tumor classification using transfer learning from deep convolutional neural networks," J. Med. Imaging, Vol.3, No.3, pp.034501, 2016. doi: 10.1117/1.JMI.3.3.034501.
[21] J. Ferlay et al., "Cancer statistics for the year 2020: An overview," Int. J. Cancer, Vol.149, No.4, pp.778–789, 2021. doi: 10.1002/ijc.33588.
[22] S. Lei et al., "Global patterns of breast cancer incidence and mortality: A population-based cancer registry data analysis from 2000 to 2020," Cancer Commun., Vol.41, No.12, pp.1183–1194, 2021. doi: 10.1002/cac2.12197.
[23] J. S. Marks et al., "Implementing recommendations for the early detection of breast and cervical cancer among low-income women," MMWR Recomm. Rep., Vol.49, pp.35–55, 2000.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors contributing to this journal agree to publish their articles under the Creative Commons Attribution 4.0 International License, allowing third parties to share their work (copy, distribute, transmit) and to adapt it, under the condition that the authors are given credit and that in the event of reuse or distribution, the terms of this license are made clear.




