A Novel Wheat Leaf Disease Classifier Leveraging Generative Adversarial Networks
DOI:
https://doi.org/10.26438/ijcse/v13i2.16Keywords:
Wheat diseases, Image classification, Deep learning, Precision agriculture, Convolutional neural networks, Generative Adversial Networks, Data AugmentationAbstract
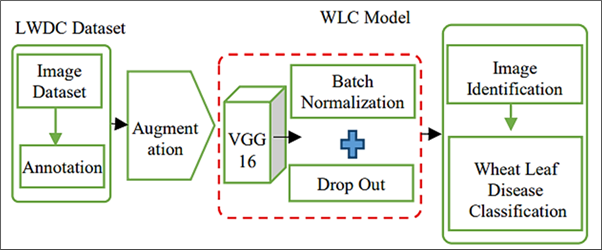
Automatic diagnosis and control of wheat plant disease are highly desired by agricultural experts. Accurate diagnosis of wheat leaf diseases is important for effective crop management. This study introduces a Wheat Leaf Convolutional (WLC) model, an enhancement of the VGG16 architecture, designed to detect and classify six distinct types of wheat leaf diseases using deep learning techniques. The model is trained using wheat leaf images dataset, augmented by Generative Adversarial Networks (GANs) to improve generalization. The WLC model got an accuracy of 94.88%, outperforming classical CNN models such as ResNet-50, AlexNet, and MobileNet by significant margins. Key metrics, including recall, precesion and F1-score, were evaluated across six disease categories: Leaf Rust, Black Chaff, Powdery Mildew, Wheat Streak, Septoria, and Healthy plants. Experimental results show that the WLC model accurately and efficiently identifies diseases, making it a useful tool for real- time applications in precision agriculture. This work contributes to improving wheat disease diagnosis, enabling timely interventions and better crop management practices.
References
[1] N. Jouini, M. Abdelkrim, and T. Ben-Hamadou, “Wheat Leaf Disease Detection: A Lightweight Approach with Shallow CNN-Based Feature Refinement”, Journal of Precision Agriculture and Machine Learning, Vol.7, Issue.2, pp.145–162, 2024.
[2] Goyal, C. M. Sharma, A. Singh, and P. K. Singh, “Leaf and Spike Wheat Disease Detection & Classification Using an Improved Deep Convolutional Architecture”, Informatics in Medicine Unlocked, Article 100642, Vol.25, 2021.
[3] M. H. Hossen, M. Mohibullah, C. S. Muzammel, T. Ahmed, S. Acharjee, and M. B. Panna, “Wheat Diseases Detection and Classification Using Convolutional Neural Network (CNN)”, International Journal of Advanced Computer Science and Applications (IJACSA), Vol.13, Issue.11, pp.719–726, 2022.
[4] M. Long, M. Hartley, R. J. Morris, and J. K. M. Brown, “Classification of Wheat Diseases Using Deep Learning Networks with Field and Glasshouse Images”, Plant Pathology, Vol.72, pp.536–547, 2023.
[5] M. A. Genaev, E. S. Skolotneva, E. I. Gultyaeva, E. A. Orlova, N. P. Bechtold, and D. A. Afonnikov, “Image-Based Wheat Fungi Diseases Identification by Deep Learning”, Plants, Vol.10, Issue.8, pp.1–21, 2021. DOI: 10.3390/plants10081500.
[6] D. Kumar and V. Kukreja, “Deep Learning in Wheat Diseases Classification: A Systematic Review”, Multimedia Tools and Applications, Vol.81, pp.10143–10187, 2022. DOI: 10.1007/s11042-022-12160-3.
[7] X. Wen, M. Zeng, J. Chen, M. Maimaiti, and Q. Liu, “Recognition of Wheat Leaf Diseases Using Lightweight Convolutional Neural Networks Against Complex Backgrounds “, Life, Vol.13, Issue.11, pp.1–22, 2023. DOI: 10.3390/life13112125.
[8] Bebronne, R., Carlier, A., Meurs, R., Leemans, V., Vermeulen, P., Dumont, B., Mercatoris, B., “In-field Proximal Sensing of Septoria Tritici Blotch, Stripe Rust, and Brown Rust in Winter Wheat by Means of Reflectance and Textural Features from Multispectral Imagery”, Biosystems Engineering, Vol.197, pp.257–269, 2020.
[9] Ficke, A., Cowger, C., Bergstrom, G., Brodal, G., “Understanding Yield Loss and Pathogen Biology to Improve Disease Management: Septoria Nodorum Blotch—A Case Study in Wheat” , Plant Disease, Vol.102, pp.696–707, 2018.
[10] Jiang, Z., Dong, Z., Jiang, W., Yang, Y., “Recognition of Rice Leaf Diseases and Wheat Leaf Diseases Based on Multi-Task Deep Transfer Learning”, Computers and Electronics in Agriculture, Article 106184, Vol.186, 2021.
[11] Too, E. C., Yujian, L., Njuki, S., Yingchun, L., “A Comparative Study of Fine-Tuning Deep Learning Models for Plant Disease Identification “, Computers and Electronics in Agriculture, Vol.161, pp.272–279, 2019.
[12] Jiang, J., Liu, H., Zhao, C., He, C., Ma, J., Cheng, T., Zhu, Y., Cao, W., Yao, X., “Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs” , Remote Sensing, Article 3446, Vol.14, Issue.14, 2022.
[13] Lin, Z., Mu, S., Huang, F., Mateen, K. A., Wang, M., Gao, W., Jia, J., “A Unified Matrix-Based Convolutional Neural Network for Fine-Grained Image Classification of Wheat Leaf Diseases” , IEEE Access, Vol. 7, pp. 11570–11590, 2019.
[14] Saleem, M. H., Potgieter, J., Arif, K. M., “Plant Disease Detection and Classification by Deep Learning” , Plants, Vol. 8, Article 468, 2019. DOI: 10.3390/plants8110468.
[15] Barbedo, J., “Impact of Dataset Size and Variety on the Effectiveness of Deep Learning and Transfer Learning for Plant Disease Classification”, Computers and Electronics in Agriculture, Vol.153, pp.46–53, 2018.
[16] Jahan, N., Flores, P., Liu, Z., Friskop, A., Mathew, J. J., Zhang, Z., “Detecting and Distinguishing Wheat Diseases Using Image Processing and Machine Learning Algorithms”, American Society of Agricultural and Biological Engineers, Article 10.13031/aim.202000372, 2020.
[17] Ransom, J. K., McMullen, M. V., “Yield and Disease Control on Hard Winter Wheat Cultivars with Foliar Fungicides”, Agronomy Journal, Vol.100, pp.1130–1137, 2008.
[18] Sharma, R. C., Nazari, K., Amanov, A., Ziyaev, Z., Jalilov, A. U., “Reduction of Winter Wheat Yield Losses Caused by Stripe Rust Through Fungicide Management”, Journal of Phytopathology, Vol.164, pp.671–677, 2016. DOI: 10.1111/jph.12490.
[19] M. Ashraf, M. Abrar, N. Qadeer, A. A. Alshdadi, T. Sabbah, and M. A. Khan, “A Convolutional Neural Network Model for Wheat Crop Disease Prediction”, Computational Materials Science, Vol.75, Issue.2, pp.3867–3882, 2023.
[20] Long, M., Hartley, M., Morris, R. J., Brown, J. K. M., “Classification of Wheat Diseases Using Deep Learning Networks with Field and Glasshouse Images”, Plant Pathology, Vol.72, Issue.3, pp.536–547, 2023.
[21] Ramadan, S. T. Y., Sakib, T., Haque, M. M. U., Sharmin, N., Rahman, M. M., “Wheat Leaf Disease Synthetic Image Generation from Limited Dataset Using GAN”, Human-Centric Smart Computing, Smart Innovation, Systems and Technologies, Vol.376, Springer, 2024.
[22] S. Sheenam, S. Khattar, and T. Verma, “Automated Wheat Plant Disease Detection Using Deep Learning: A Multi-Class Classification Approach”, 2023 3rd International Conference on Intelligent Technologies (CONIT), Hubli, India, pp.1–5, 2023.
[23] Yakkundimath, R., Saunshi, G., Anami, B., “Classification of Rice Diseases Using Convolutional Neural Network Models”, Journal of the Institution of Engineers (India) Series B, Vol.103, pp.1047–1059, 2022. DOI: 10.1007/s40031-021-00704-4.
[24] Yakkundimath, R., Saunshi, G., Kamatar, V., “Plant Disease Detection Using IoT” , International Journal of Engineering Science and Computing, Vol.8, Issue.9, pp.18902–18906, 2018.
[25] Malvade, N. N., Yakkundimath, R., Saunshi, G., Elemmi, M. C., “A Comparative Analysis of Paddy Crop Biotic Stress Classification Using Pre-Trained Deep Neural Networks”, Artificial Intelligence in Agriculture, Vol.6, pp.167–175, 2022. DOI: 10.1016/j.aiia.2022.09.001.
[26] Yakkundimath, R., Saunshi, G., Palaiah, S., “Automatic Methods for Classification of Visual-Based Viral and Bacterial Disease Symptoms in Plants “, International Journal of Information Technology, Vol.14, pp.287–299, 2022. DOI: 10.1007/s41870-021-00701-2.
[27] Yakkundimath, R., Saunshi, G., “Identification of Paddy Blast Disease Field Images Using Multi-Layer CNN Models”, Environmental Monitoring and Assessment, Vol.195, Article 646, 2023. DOI: 10.1007/s10661-023-11252-3.
[28] Malvade, N. N., Yakkundimath, R., Saunshi, G., Elemmi, M. C., Baraki, P., “Paddy Variety Identification from Field Crop Images Using Deep Learning Techniques”, International Journal of Computational Vision and Robotics, Vol.13, Issue.4, pp.405–419, 2023. DOI: 10.1504/IJCVR.2023.131986.
[29] Saunshi, G., Chini, S., Ganvatkar, P., Nayak, R., “Identification and Classification of Medicinal Leaves and Their Medicinal Values”, 2023 4th IEEE Global Conference for Advancement in Technology (GCAT), Bangalore, India, pp.1–4, 2023. DOI: 10.1109/GCAT59970.2023.10353283.
[30] S. Sajjan, G. Saunshi, and S. Hiremath, “Contour-Based Leaf Segmentation in Green Plant Images”, 2022 2nd Asian Conference on Innovation in Technology (ASIANCON), Ravet, India, pp.1–5, 2022. DOI: 10.1109/ASIANCON55314.2022.9909217.
[31] Badiger, R. M., Yakkundimath, R., Konnurmath, G., and Dhulavvagol, P. M., “Deep Learning Approaches for Age-Based Gesture Classification in South Indian Sign Language”, Engineering, Technology & Applied Science Research, Vol.14, Issue 2, pp. 13255–13260, 2024. DOI: 10.48084/etasr.6864.
[32] R. Yakkundimath, R. Badiger, and N. Malvade, “Deep Learning-Based Classification of Single-Hand South Indian Sign Language Gestures”, Journal of Electrical Systems, Vol. 20, Issue 2s, pp. 200–209, 2024.
[33] Badiger, R. M., Y. K. Manjunath, and T. P. Nigappa, “Content Extraction from Advertisement Display Boards Utilizing Region Growing Algorithm”, International Journal of Computer Vision, Vol.45, Issue.2, pp.123–134, 2016.
[34] Badiger, R. M., and D. Lamani, “Deep Learning-Based South Indian Sign Language Recognition by Stacked Autoencoder Model and Ensemble Classifier on Still Images and Videos”, Journal of Theoretical and Applied Information Technology, Vol.100, Issue.21, pp.6587–6597, 2022.
[35] Badiger, R. M., and D. Lamani, “Recognition of South Indian Sign Languages for Still Images Using Convolutional Neural Network”, International Journal of Future Generation Communication and Networking, Vol.14, Issue.1, pp.832–843, 2021.
[36] N. N. Malvade, R. Yakkundimath, G. B. Saunshi, and M. C. Elemmi, “Paddy Variety Identification from Field Crop Images Using Deep Learning Techniques”, International Journal of Computational Vision and Robotics, Vol.13, Issue.4, pp.405–419, July 2023. DOI: 10.1504/IJCVR.2023.131986.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors contributing to this journal agree to publish their articles under the Creative Commons Attribution 4.0 International License, allowing third parties to share their work (copy, distribute, transmit) and to adapt it, under the condition that the authors are given credit and that in the event of reuse or distribution, the terms of this license are made clear.




